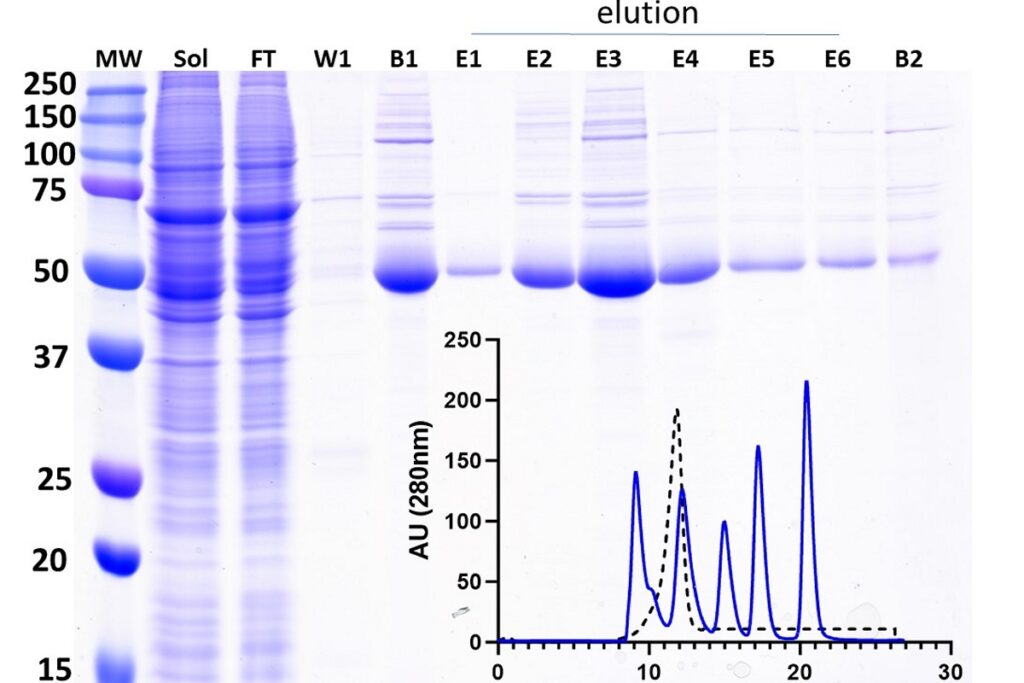
The HTSC Core provides scaled expression of recombinant proteins to the Penn research community. Proteins can be used for biophysical/biochemical characterization, crystalography, cryo-EM, and high-throughput screening to identify small molecule inhibitors. The HTSC Core offers services to express proteins in E. coli, baculovirus expression vector systems, and mammalian cells using the Expi293 system. These activities are provided through a centralized laboratory under the direction of an experienced Core Director who has a history of collaboration with investigators on protein expression projects. The Core supports all aspects of recombinant protein expression, including expression construct design, baculovirus preparation, and analytical/ preparative scale productions in all expression systems. The Core also provides protein purification services. Protocols developed by the core are used to purify recombinant proteins from soluble extracts or cell supernatants, using appropriate affinity matrices. When necessary, a second purification step (e.g., affinity, ion exchange, size exclusion chromatography, etc.) will be used to ensure maximum purity of the recombinant protein.
For more information regarding these services, contact David C. Schultz, Ph.D at dschultz@pennmedicine.upenn.edu.



