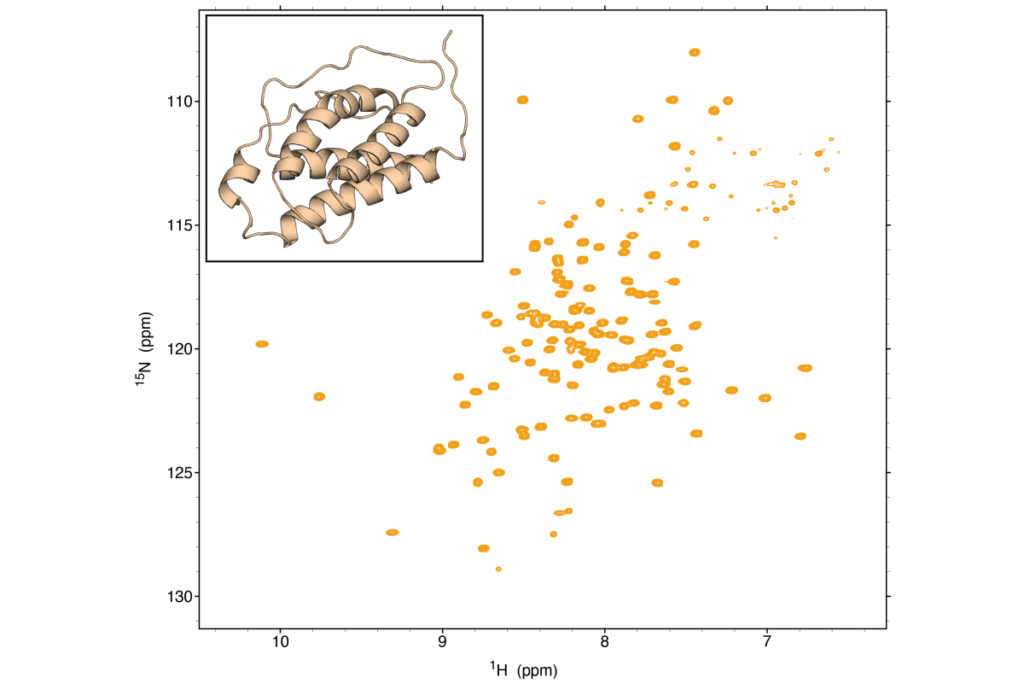
We utilize NMR spectroscopy for studying the structures, dynamics, and interactions of proteins and nucleic acids. NMR is essential technique for understanding protein-ligand interactions, protein-protein interactions, and conformational changes. NMR is also uniquely capable to elucidate structures and conformational dynamics of RNA, RNA-protein and RNA-small molecule interactions. Obtaining insights into complex interactions, characterizing biomolecular dynamics, and determining pathways for conformational exchange not only reveals insights into biological function, but provides molecular blueprints to develop new therapeutics.
Shared facility
https://www.med.upenn.edu/jf/bsbcore/index.html
The biomolecular NMR facility allows studies of proteins, RNA and other important biomolecules in a physiologically relevant, aqueous environment. Our facility is equipped with 600 MHz and 800 MHz Avance Neo NMR systems with 5mm 1H-optimized triple resonance cryoprobe designed for 1H or 19F observation with 13C/15N decoupling, and 13C observation with 1H decoupling due to superior sensitivity on 13C. These capabilities are utilized in structure determination, studies of intrinsically disordered proteins, ligand binding and fragment-based drug discovery applications which serve the broader structural biology community at Penn and CHOP.




